pyaibox.ml package
Submodules
pyaibox.ml.dataset_visualization module
- pyaibox.ml.dataset_visualization.visds(x, y=None, labelmap=None, axisn=0, mode='bar(nms)', **kwargs)
pyaibox.ml.reduction_pca module
- pyaibox.ml.reduction_pca.pca(x, axisn=0, ncmpnts='auto99%', algo='svd')
Principal Component Analysis (pca) on raw data
- Parameters
- Returns
U, S, K (if
ncmpntsis integer)- Return type
array
Examples

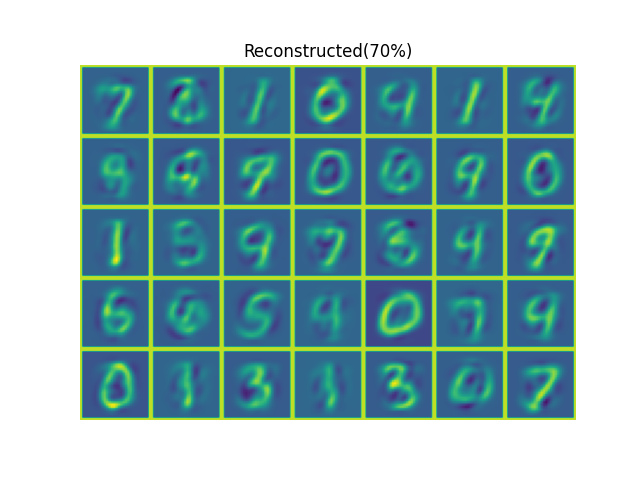
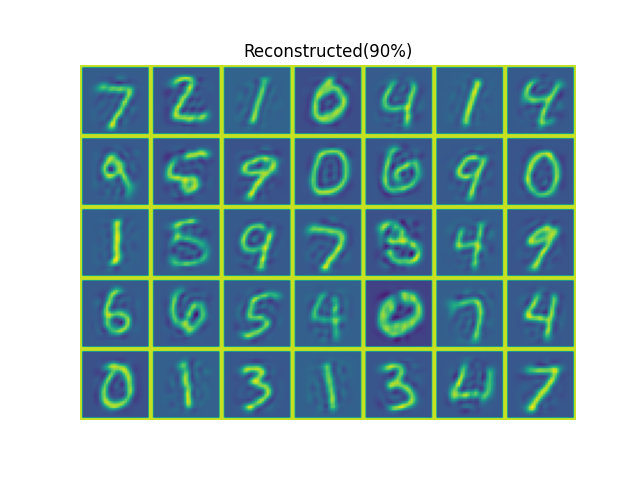
The results shown in the above figure can be obtained by the following codes.
import numpy as np import pyaibox as pb rootdir, dataset = '/mnt/d/DataSets/oi/dgi/mnist/official/', 'test' x, _ = pb.read_mnist(rootdir=rootdir, dataset=dataset, fmt='ubyte') print(x.shape) N, M2, _ = x.shape u, s, k = pb.pca(x, axisn=0, ncmpnts='auto90%', algo='svd') print(u.shape, s.shape, k) u = u[:, :k] y = x.reshape(N, -1) @ u # N-k z = y @ u.T.conj() z = z.reshape(N, M2, M2) print(pb.nmse(x, z, axis=(1, 2))) xp = np.pad(x[:35], ((0, 0), (1, 1), (1, 1)), 'constant', constant_values=(255, 255)) zp = np.pad(z[:35], ((0, 0), (1, 1), (1, 1)), 'constant', constant_values=(255, 255)) plt = pb.imshow(pb.patch2tensor(xp, (5*(M2+2), 7*(M2+2)), axis=(1, 2)), titles=['Orignal']) plt = pb.imshow(pb.patch2tensor(zp, (5*(M2+2), 7*(M2+2)), axis=(1, 2)), titles=['Reconstructed' + '(90%)']) u, s, k = pb.pca(x, axisn=0, ncmpnts='auto0.7', algo='svd') print(u.shape, s.shape, k) u = u[:, :k] y = x.reshape(N, -1) @ u # N-k z = y @ u.T.conj() z = z.reshape(N, M2, M2) print(pb.nmse(x, z, axis=(1, 2))) zp = np.pad(z[:35], ((0, 0), (1, 1), (1, 1)), 'constant', constant_values=(255, 255)) plt = pb.imshow(pb.patch2tensor(zp, (5*(M2+2), 7*(M2+2)), axis=(1, 2)), titles=['Reconstructed' + '(70%)']) plt.show() u, s = pb.pca(x, axisn=0, ncmpnts=2, algo='svd') print(u.shape, s.shape) y = x.reshape(N, -1) @ u # N-k z = y @ u.T.conj() z = z.reshape(N, M2, M2) print(pb.nmse(x, z, axis=(1, 2)))